
You may use the grep function with the following arguments to eliminate a small number of genes from a bigger list: Taking one list and subtracting it from another:

To count the number of sequences in AT cDNA.fa and RefSeq.faa, we apply the following strategy:Ģ. However, if the deflines include > more than once, the count will be messed up! You can utilize the following to be safe: This may be done using the grep count option -c and the counting option of the grep. You may count the number of sequences by counting > in the file. The number of sequences in a file should be equal to the number of description lines, according to the FASTA format standard. Let’s now use a DNA program to practical illustrate how the grep command is used to do some basic tasks on several sequences: It also provides support for regular expressions in pattern matching and can highlight the context of a match by displaying lines before and after the result.
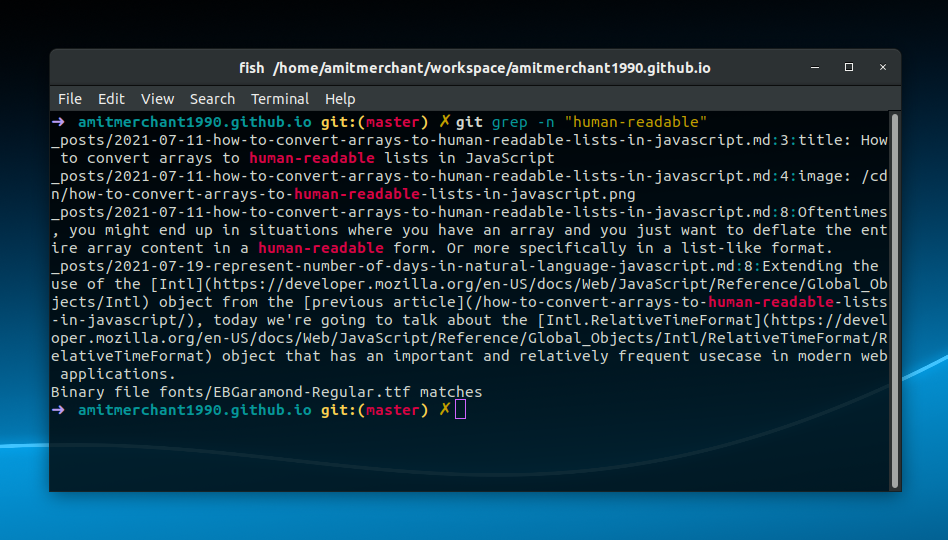
Look for restriction spots in the sequence (s).In a sequence file, look for a matching theme.Get the fasta sequence file’s header lines.The number of sequences in a multi-fasta sequence file may be counted.The following is a list of some of the scenarios in which you might want to use grep: grep can be used on its own or in conjunction with pipes.Įxample when pipe is used: $ ls -l | grep rwxrwxrwxĮxample when pipe isn’t used: $ grep -r "192.168.1.5" /etc/

It may be used to recursively search a directory structure of files for text and to find text in a file. In UNIX, the grep command is used to output lines that match a pattern. Grep is an acronym that stands for “global regular expression printer.” In layman’s terms, grep retrieves the lines from a supplied text that fit the user’s criteria. Grep is by far the most often used command in Unix.


 0 kommentar(er)
0 kommentar(er)
